phylogenetic tree
 LUCA ultimately gave rise to the prokaryotic Bacteria (Eubacteria) and the Archaea.
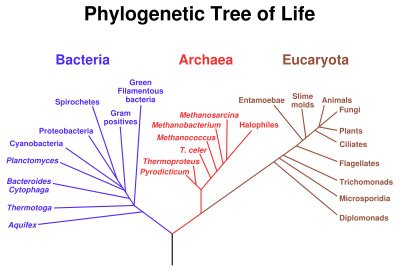
LUCA ultimately gave rise to the prokaryotic Bacteria (Eubacteria) and the Archaea.Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant phenotypical (phenetic) taxonomies of the prokaryotes. Phenetic systems group organisms based on mutual similarity of phenotypic (physical and chemical) characteristics. Phenetic groupings may or may not correlate with evolutionary relationships. The image above left shows a phylogenetic tree of living things, based on 16s rRNA data and proposed by Carl Woese, showing the separation of bacteria, archaea, and eukaryotes.
Trees constructed with other genes are generally similar to the 16s rRNA tree, although they may place some early-branching groups very differently due to analysis problems arising from long branch attraction. The exact relationships of the three domains are still being debated, as is the position of the root of the tree. It has also been suggested that due to horizontal (lateral) gene transfer, a tree may not be the best representation of the genetic relationships of all organisms.
Long branch attraction (LBA) is a problem in phylogenetic analyses, particularly in those analyses employing the non-parametric statistical method termed maximum parsimony. In LBA, rapidly evolving lineages are inferred to be closely related, regardless of their true evolutionary relationships. This problem in analysis arises when the DNA of two (or more) lineages evolves rapidly. Because there are only four possible nucleotides, high rates of DNA substitution create the probability that two separate lineages will convergently evolve the same nucleotide at the same locus. In such cases, parsimony erroneously interprets this similarity as a synapomorphy, that is, as having evolved once in the common ancestor of the two lineages. The problem of LBA can be minimized by applying statistical methods that incorporate differential rates of substitution among lineages, such as maximum likelihood, or by breaking up long branches by adding taxa that are related to those with the long branches.
A synapomorphy is a derived character-state that is shared by two or more terminal groups – taxa included in cladistic analyses as being units that cannot be further divided. The pertinent character-states in synapomorphy are inherited from their most recent common ancestor.
αΩ Beginning αΩ Geological Time ▫ site map
tags [Evolution] [phylogenetic tree] [evolutionary relationship] [clade] [phenetic] [taxonomy] [domains of life]
Labels: cladistics, evolutionary relationships, phenetic, phylogenetic tree







































